IMPETUS – Interactive MultiPhysics EnvironmenT for Unified Simulations
an object oriented, easy-to-use, high performance, C++ program for three-dimensional simulations of complex physical systems that can benefit a large variety of research areas, especially in cell mechanics. The program implements cross-communication between locally interacting particles and continuum models residing in the same physical space while a network facilitates long-range particle interactions. Message Passing Interface is used for inter-processor communication for all simulations.
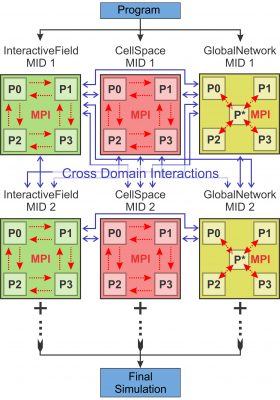 IMPETUS Design Flow
IMPETUS Design Flow
The diagram illustrates the blueprint of how a typical IMPETUS model is constructed. Individually, each Modular Interactive Domain (MID) is a simulation by itself. By assembling different MID components together, users can create a large number of different simulation models. IMPETUS provides a large variety of tools for users to easily design and program the cross-domain interactions.
IMPETUS VR – Immersive Virtual Reality Environment for IMPETUS Simulations
Simulations - Long & Short Format
IMPETUS VR is an interactive virtual reality environment for particle dynamics simulations using IMPETUS. This is a tool using immersive interactive virtual reality technology to assist users to develop molecular dynamics simulations by placing the user in the live simulation world to interact with and reconfigure the particles.
MD-R - Molecular Dynamics library for Repast
The MD-R toolkit provides a three-dimensional continuous space projection and a large set of short-range molecular dynamics (MD) tools for the Agent-based modeling toolkit Repast HPC.
The 3D projection gives Repast HPC the ability to simulate agents in three dimensional Cartesian space and it manages all MPI operations for parallel computing. The MD-R toolkit utilizes the provided 3D space projection to run short-range Molecular Dynamics simulations. It includes integration algorithms, thermostats, and potential functions for interacting agents. MD-R was designed to be intuitive and user friendly. Users are not required to have advanced knowledge of MD to create a simulation with MD-R. The toolkit automatically handles all major MD operations such as initiation, recording of MD parameters, creation of cell lists and neighbor lists containing the agents. It also determines when the agents need to be synchronized and applies thermodynamic constrains.
Three-dimensional continuous space projection for Repast
MD-R - 3D Projection - The Three-dimensional continuous space projection for Repast HPC is available to download here (link not available)
A demo to run the 3D space is available to download here (link not available)
- Instruction for the demo is located in the 'readme' folder in the demo and can be viewed here: txt , pdf (link not available)
- This 3D demo model uses AtomEye for molecular visualization. Please download AtomEye to view the motion of agents. (link not available)
Molecular Dynamics library for Repast
The Molecular Dynamics library for Repast is available to download here (link not available)
A demo to run a molecular dynamics library for repast is available to download here (link not available)
- Instruction for the demo is located in the 'readme' folder in the demo and can be viewed here: txt , pdf (link not available)
- The template files are located in the folder mdr-templates in the demo folder or it can be downloaded from here (link not available)
- This demo model uses AtomEye for molecular visualization. Please download AtomEye to view the motion of agents (link not available)
FRAME - Force Review Automation Environment
We have created an open-source software environment using MATLAB, the Force Review Automation Environment (FRAME), to greatly increase the speed at which AFM data are processed. FRAME features a number of algorithms to streamline automated processing as well as a GUI interface to expedite manual processing and review of the data. The results can be displayed in FRAME with MATLAB plotting tools, as well as exported as a Microsoft Excel file, for further statistical analysis. FRAME is meant to be easy to use, even for the novice MATLAB user.
FRAME in Video
Additional Software
This is an in-house developped set of MATLAB functions useful for counting and quantifying the morphology of red blood cells from light microscopy videos.